Understanding
mitotic nuclear remodeling from the comparative and synthetic
biology perspectives constitutes the main thrust of our
current research.
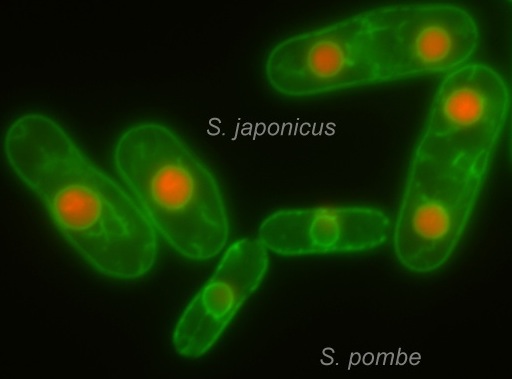
We use two fission yeast species, Schizosaccharomyces pombe and Schizosaccharomyces japonicus that employ strikingly divergent mitotic strategies to address the following questions:
How
is the nuclear envelope area controlled during mitotic
division?
How
do cells break and reform the nuclear membrane?
What
are the rules for generating functional diversity in nuclear
envelope management?
Aside from the nuclear envelope biology, the two fission yeasts exhibit intriguing differences in several integral biological processes including cytokinesis, cell size control, chromatin organization, and polarized growth. We also use S. japonicus and S. pombe to understand the interplay between cellular division site placement and cortical patterning. We have been developing new avenues of research aimed at understanding the mechanisms underlying differences in overall cell size and cell size homeostasis in the two sister species and the scaling of chromosome compaction during mitosis.
Copyright © 2016 by Snezhana Oliferenko